Kvik
A framework for developing interactive data exploration applications in genomics
and systems biology. It is a collection of packages in the Go programming
language that help application developers i) interface with
the R statistical programming language; ii) specify and run statistical analysis
pipelines; and iii) interface with online databases such as
MsigDB and
KEGG. It also contains a Javascript package for
visualizing KEGG pathways using d3.
Kvik is in continuous development.
Example Applications
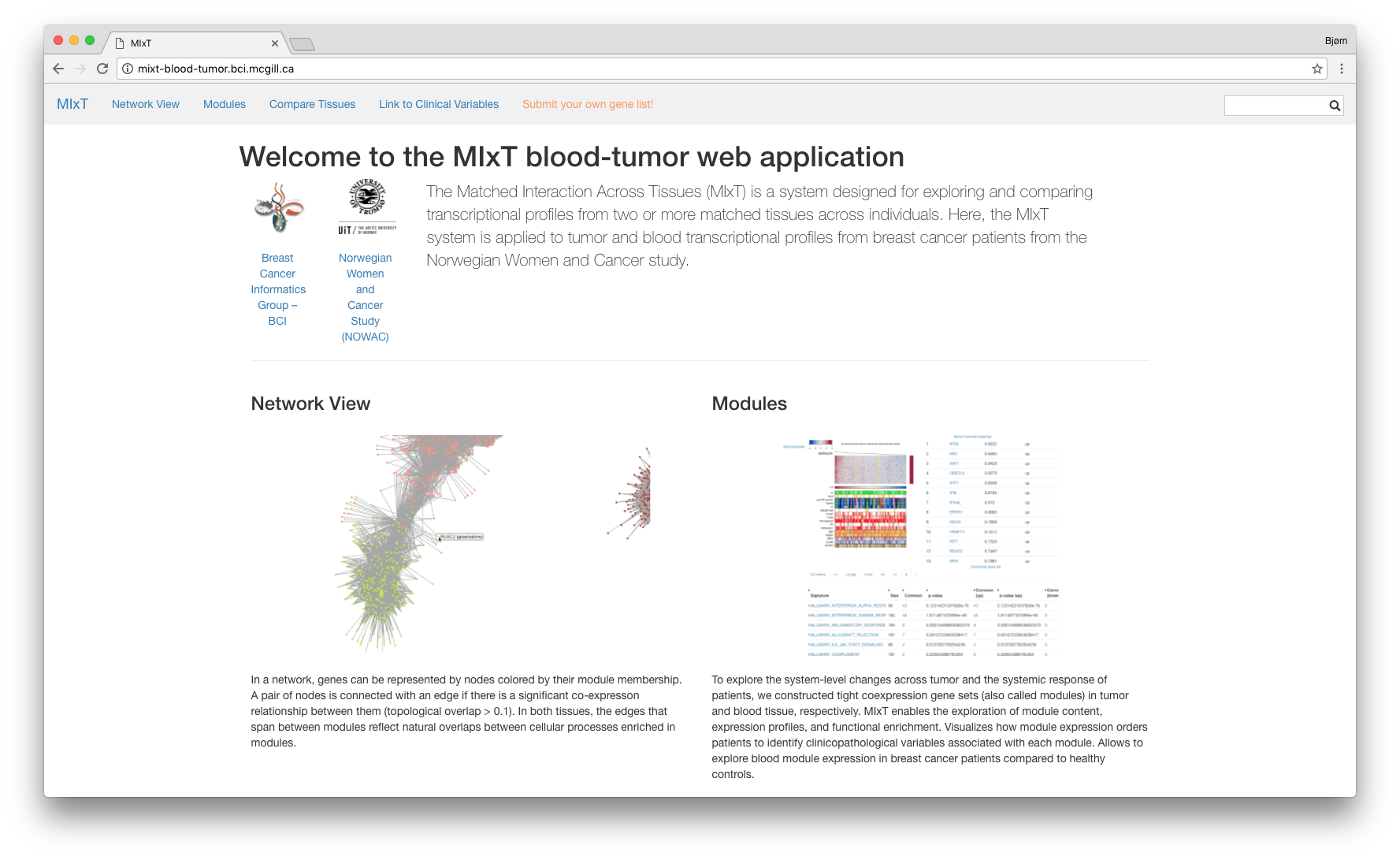
Kvik Pathways

Name
The project is named after Fridtjof
Nansen's lead slead dog Kvik
on his Fram expedition to the North Pole 1893-1896.
History
The project started as a system for exploring KEGG pathway maps together with
gene expression datasets from the Norwegian Women and Cancer
(NOWAC). Over the course of the last two years
the project has moved from being a specific application into a generic framework
for building applications that interface with data analysis in R and online
databases.
Feel free to contact me at bjorn.fjukstad@uit.no!
 Directories
¶
Directories
¶

